|
The Graphical Query Language: |
Tutorial GQLQuery
Overview
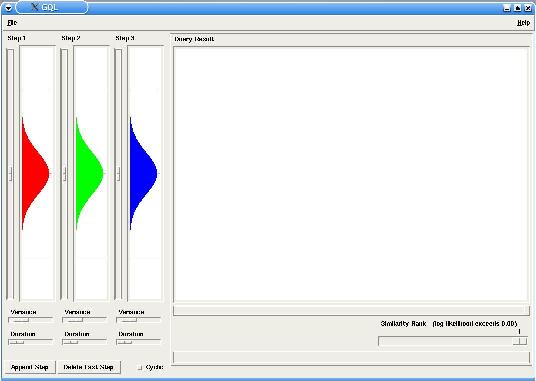
GQLQuery is a graphical interface for querying time course data given a HMM model. The interface is divided in two main parts: the Model panel in the left, where you can see and modify the model's parameters and the Query panel, where information of the queried time courses is displayed.
Getting Started
Run GQLQuery with the following command (or by double-clicking at the executable binary files):
python GQLQuery.py
After running GQLQuery, open the file yspo_2fold.txt with the Menu File -> Open Data Set. You will see all available time courses plotted in the Query panel. Next, open the sample model yspo_upregulated.smo in the Menu File -> Open Query. The Model panel is updated to reflect the loaded model parameters (see Documentation for file formats).
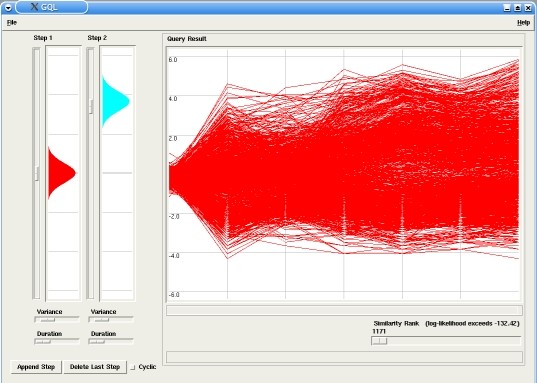
Making a Query
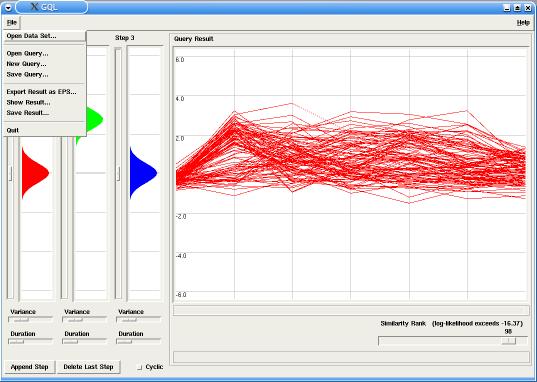
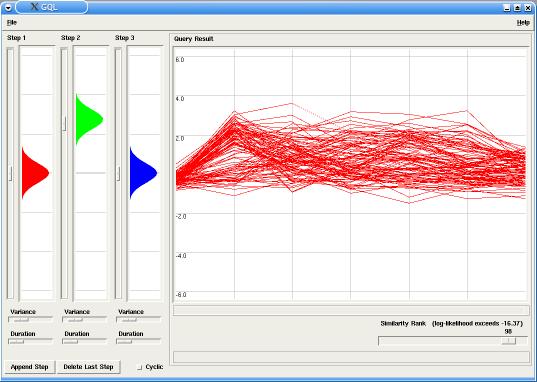
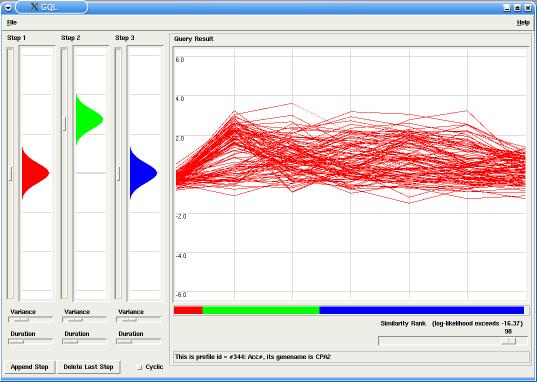
In our example the model has two steps (or states), with probabilities emissions around 0.0 for the first step and around 3.0 for the second step. By changing the similarity rank, using the corresponding slider in the Query panel, only the best fitting time-courses are displayed in the Graph (in this case, up-regulated genes).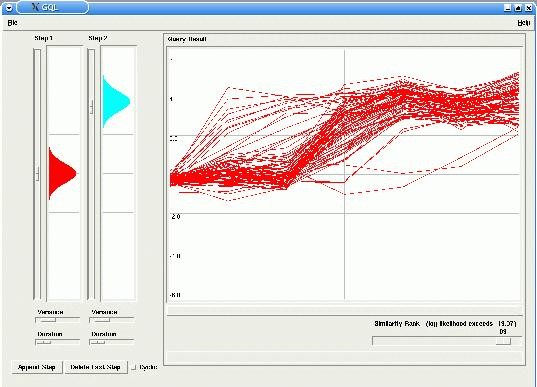


Saving your Results
After all analysis is done, all results can be saved. Through the Menu File the user can: save the current model, save the current plot in a eps format or create a data file containing only the queried time-courses.